TIG stack on G5K
Experiment Artifacts
The artifacts repository contains the E2Clab configuration files such as
layers_services.yaml, network.yaml, and workflow.yaml
$ cd ~/git/
$ git clone https://gitlab.inria.fr/E2Clab/examples/monitoring-tig-g5k.git
$ cd monitoring-tig-g5k/
Defining the Experimental Environment
Layers & Services Configuration
The monitoring type is tig with the provider (machine hosting InfluxDB and
Grafana) on the g5k testbed (paravance cluster). The network is shared
(experiment data and monitoring data are on the same network). We use an IPv4
network. Finally, we added roles: [monitoring] in all services (e.g., MyServer,
MyClientA, and MyClientB) for tig monitoring.
1---
2environment:
3 job_name: monitoring-tig-g5k
4 walltime: "00:59:00"
5 g5k:
6 cluster: paravance
7 job_type: ["allow_classic_ssh"]
8monitoring:
9 type: tig
10 provider: g5k
11 cluster: paravance
12 network: shared
13 ipv: 4
14layers:
15- name: cloud
16 services:
17 - name: MyServer
18 environment: g5k
19 cluster: paravance
20 roles: [monitoring]
21 quantity: 1
22- name: edge
23 services:
24 - name: MyClientA
25 environment: g5k
26 cluster: paravance
27 roles: [monitoring]
28 quantity: 2
29 - name: MyClientB
30 environment: g5k
31 cluster: paravance
32 roles: [monitoring]
33 quantity: 1
Network Configuration
1networks:
2- src: cloud
3 dst: edge
4 delay: 180ms
5 rate: 1gbit
6 loss: 0.1%
Workflow Configuration
prepare installs stress on all Services.
launch runs stress on all Services.
1# SERVER
2- hosts: cloud.*
3 prepare:
4 - shell: apt install -y stress
5 launch:
6 - debug:
7 msg: "Running"
8 - shell: stress --cpu 32 --timeout 60
9 async: 60
10 poll: 0
11# CLIENT
12- hosts: edge.*
13 prepare:
14 - shell: apt install -y stress
15 launch:
16 - debug:
17 msg: "Running"
18 - shell: stress --cpu 32 --timeout 60
19 async: 60
20 poll: 0
Running & Verifying Experiment Results
Find below the commands to run this example.
$ e2clab layers-services ~/git/monitoring-tig-g5k/
~/git/monitoring-tig-g5k/
$ e2clab workflow ~/git/monitoring-tig-g5k/ prepare
$ e2clab workflow ~/git/monitoring-tig-g5k/ launch
You can access the Grafana service to visualize monitoring data during experiment
execution. You can access it as described in the following file
~/git/monitoring-tig-g5k/20231013-150327/layers_services-validate.yaml. See
more details below:
'* * * * * * * Monitoring Service (started during workflow ''launch'' step)'
'Available at: http://localhost:3000'
'Access from your local machine: ssh -NL 3000:localhost:3000 paravance-10.rennes.grid5000.fr'
'username: admin / password: admin'
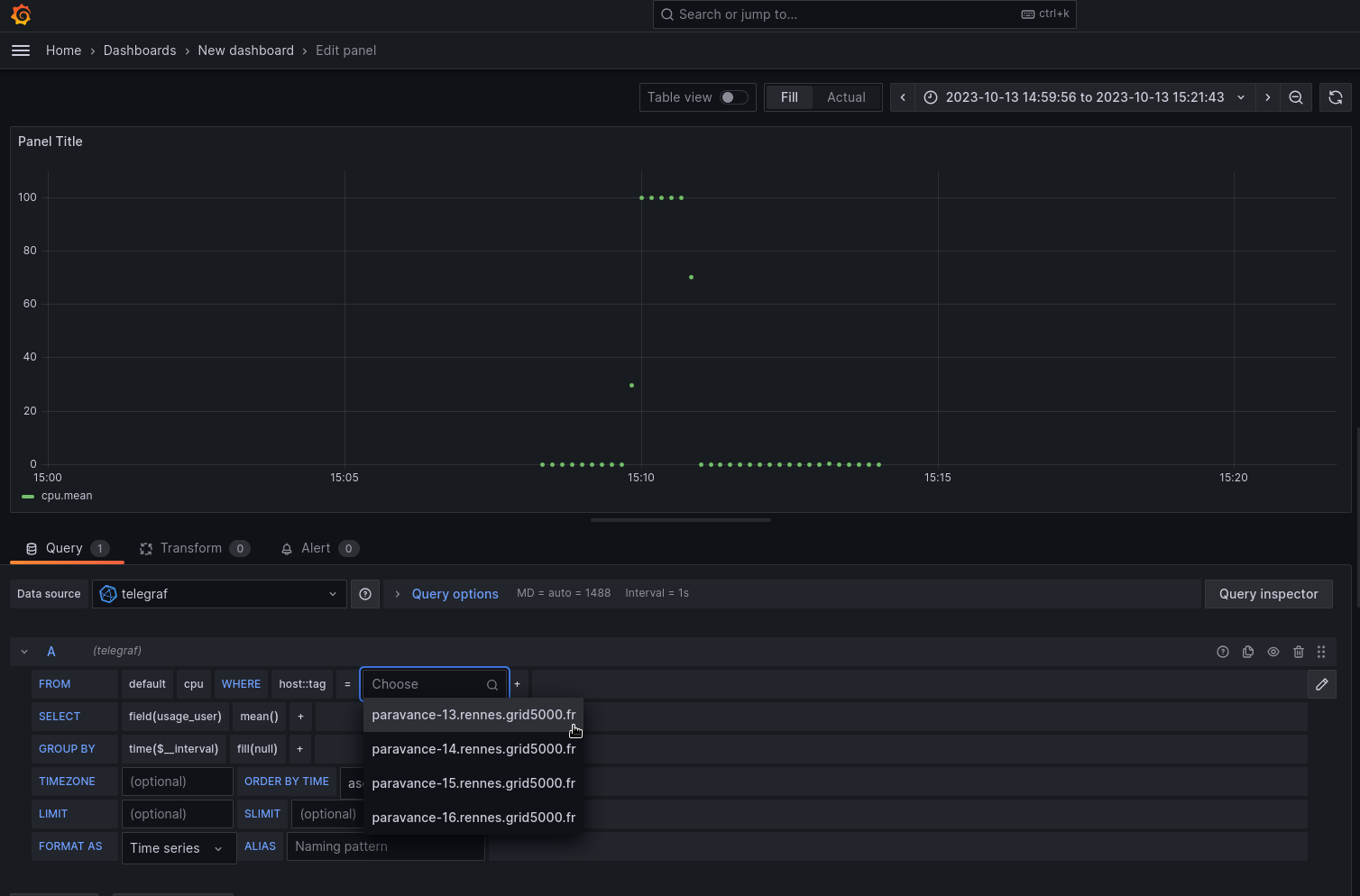
Figure 1: CPU usage on G5K node ‘MyServer’.
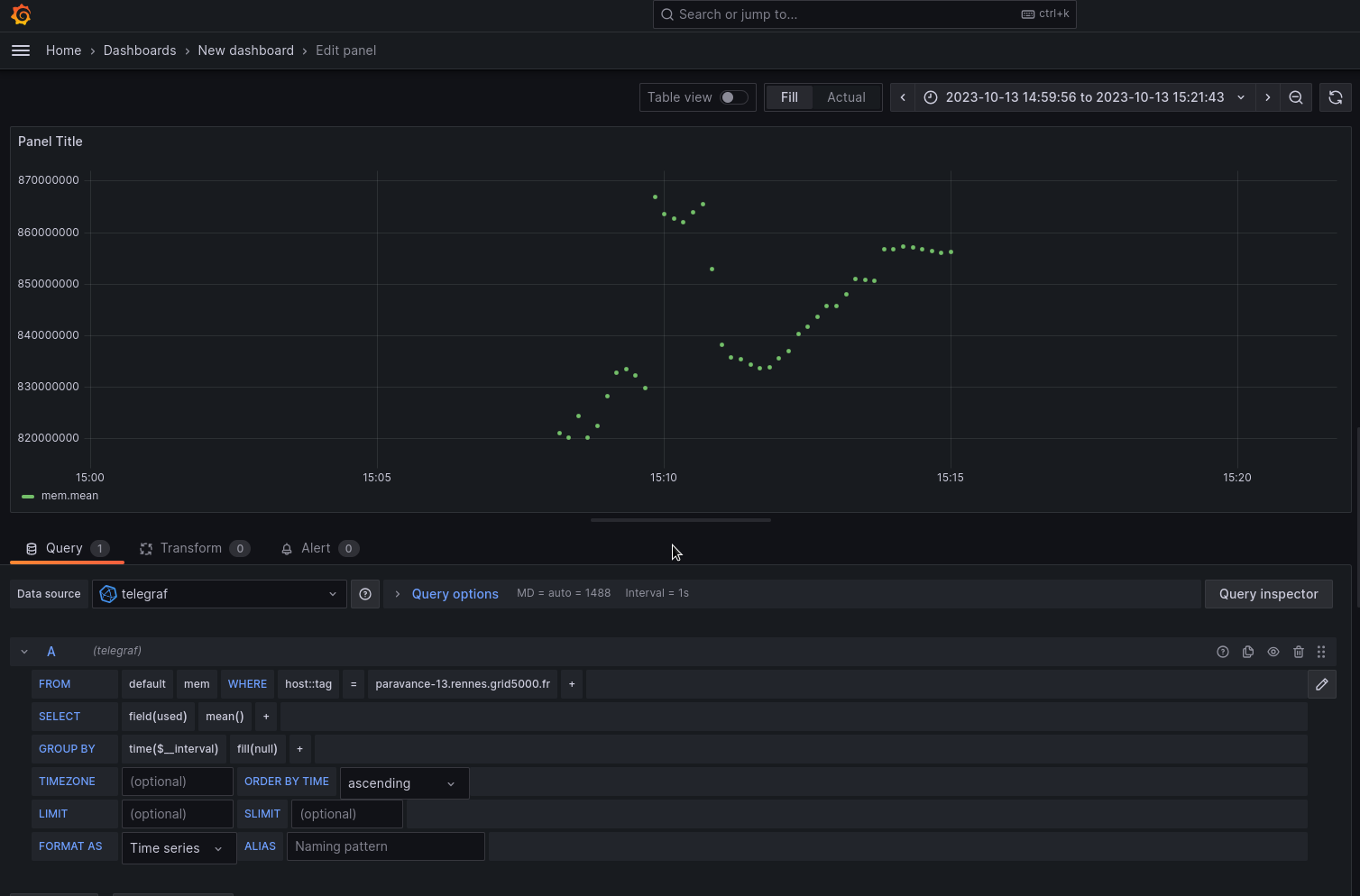
Figure 2: Memory usage on G5K node ‘MyServer’.
Wait at least one minute before finalizing the workflow and saving the monitoring data.
$ e2clab finalize ~/git/monitoring-tig-g5k/
The monitoring data will be saved at:
$ ls ~/git/monitoring-tig-g5k/20231013-150327/monitoring-data/
influxdb-data.tar.gz